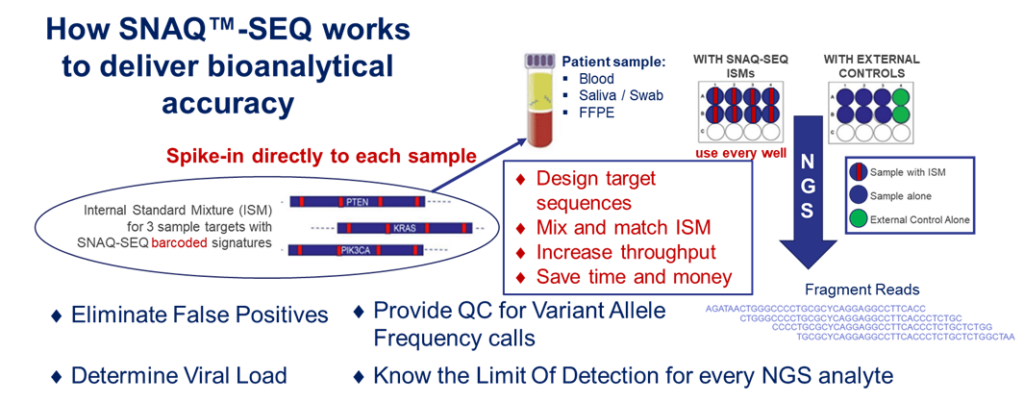
AccuGenomics’ patented and propriety technology, Standardized Nucleic Acid Quantification (SNAQ™) for Sequencing (SNAQ-SEQ) is used to manufacture Accukits™ that contain internal standard mixtures (ISM™) designed for a given set of genetic targets. These SNAQ-SEQ Spike-in standards are added to each specimen at the library prep stage of the workflow. Biochemically identical to the target regions except for the presence of a barcode (single base mutation) every 50-80 bp, each Internal Standard in the ISM will co-vary with their targets and allow measurement of background error rates due to technical or systematic errors known to occur throughout the sequencing process. The barcode signature contained within the sequence allows separate pipeline analysis of the target and ISM sequences.
What is SNAQ™-SEQ?
Standardized Nucleic Acid Quantification for SEQuencing (SNAQ-SEQ) is an innovative method that utilizes synthetic DNA or RNA internal standards that are spiked into each sample prior to extraction or NGS library preparation steps. Internal standards (standards that are added directly to the sample) are the most optimal way of controlling for variation within the result and are the gold standard method for sensitive bioanalyte measurements.
These standards will co-vary with the sample targets to provide independent biochemical QC for each sample measurement.
SNAQ-SEQ spike-in controls for NGS are compatible with amplicon and capture library preparation methods and can be run on any next generation sequencer including Illumina and Ion Torrent platforms.
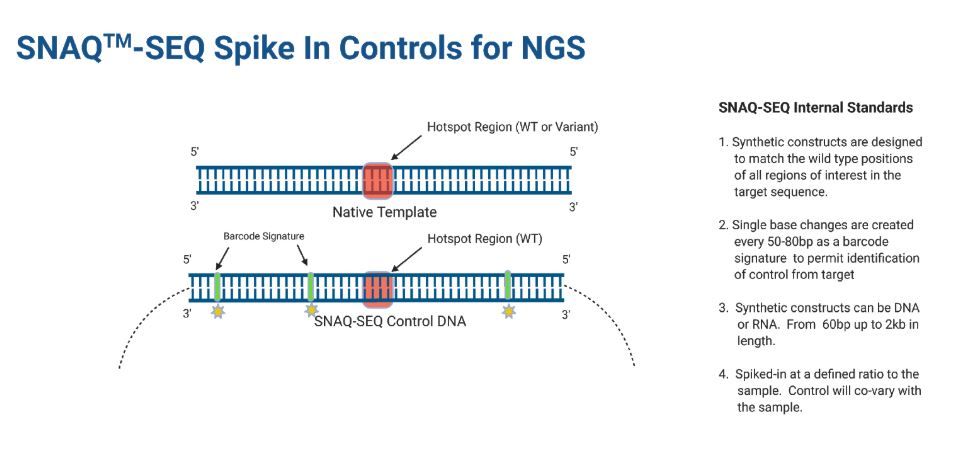
SNAQ-SEQ internal standards (IS) are constructed using synthetic DNA or RNA matching the target regions of interest and are produced using ultra-low error rate methods. The standards are resolved bioinformatically from native template (NT) using barcode signatures in which single base changes every 50-80 bp are incorporated into the sequence.
SNAQ-SEQ designs are typically to the reference sequence and contain wild type alleles at all targeted positions. Internal standards Mixtures (ISM™) are built for all actionable variants/gene regions including for SNVs, Insertions, Deletions, Fusions and CNVs. SNAQ-SEQ standards can range in size from 100bp to 2000 bp.
How does SNAQ-SEQ Work?
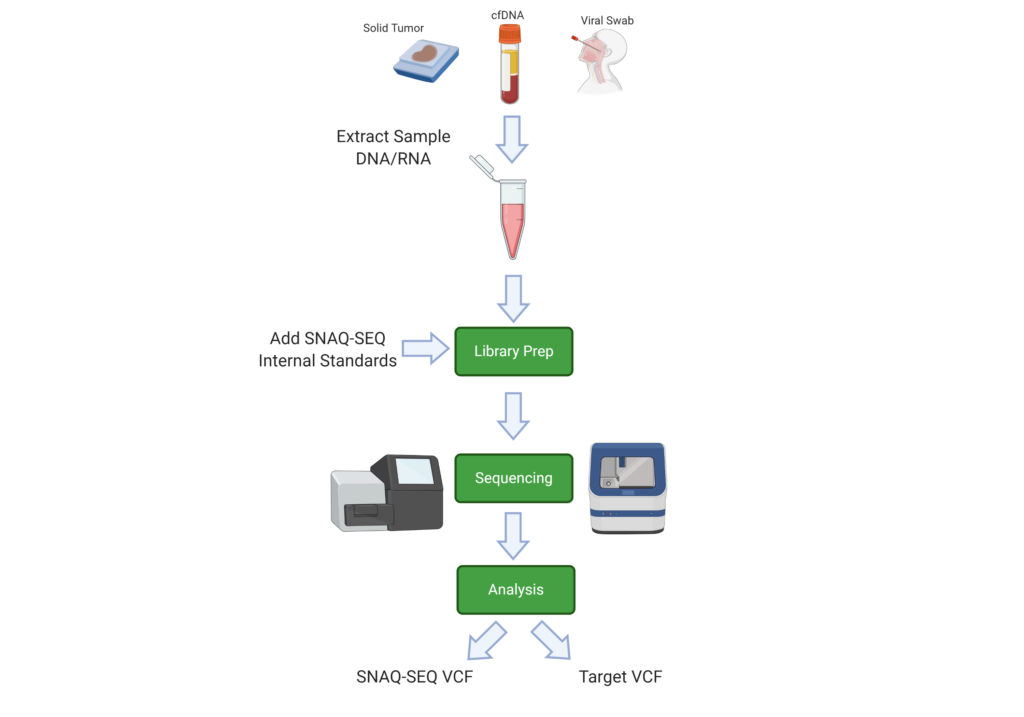
SNAQ-SEQ Spike-in standards are mixed into ISM™ and added to the specimen at the library prep stage of the workflow. Being biochemically identical to the target regions, ISM will co-vary with the target and act to measure the background error rates due to technical and systemic errors through the sequencing process. The barcode signature contained within the sequence allows separate pipeline analysis of the target and ISM sequences.
SNAQ-SEQ improves variant calling accuracy
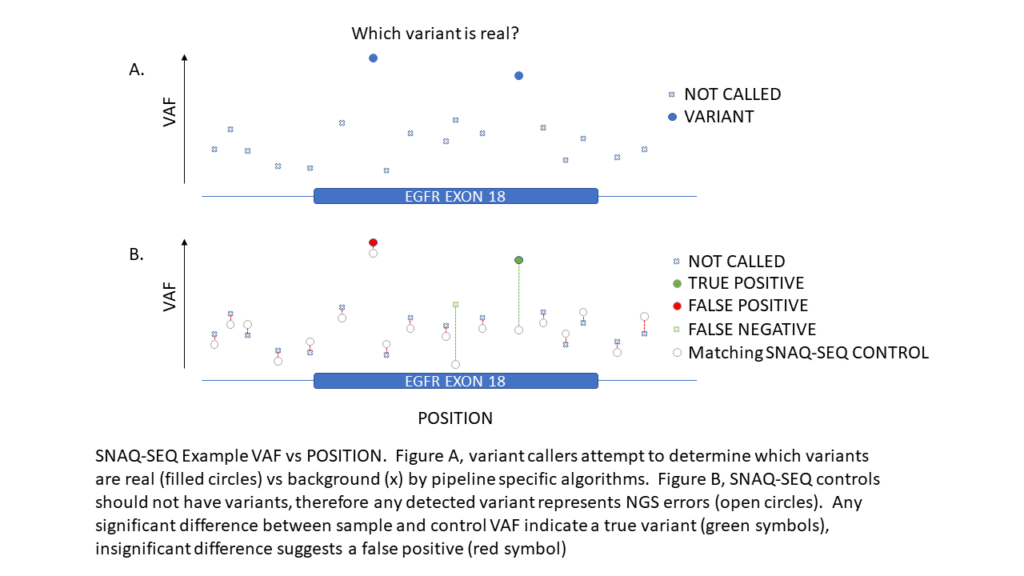
SNAQ-SEQ internal standards are identical to the native target at all controlled positions and enable highly accurate measurement of background error rates. SNAQ-SEQ provides limit of blank measurement for EACH region in EACH specimen which dramatically improves variant caller accuracy—particularly at low variant allele frequencies that are often subject to high noise and background.
Benefits of SNAQ-SEQ Spike-in standards:
- Higher sensitivity: Provides positional LOB and LOD on a PER SAMPLE basis
- More accurate calls: Measurement of background error rates due to library prep, sequencing or variant caller errors
- Elimination of False Positives for variant calls at or below LOB
- Superior CNV resolution (including LOH and low copy amplifications)
- More accurate RNASeq transcript abundance measurements with significantly less read depth
Contact AccuGenomics to see how SNAQ-SEQ standards can help your assay today!
