SNAQ-SEQ ctDNA spike-in standards
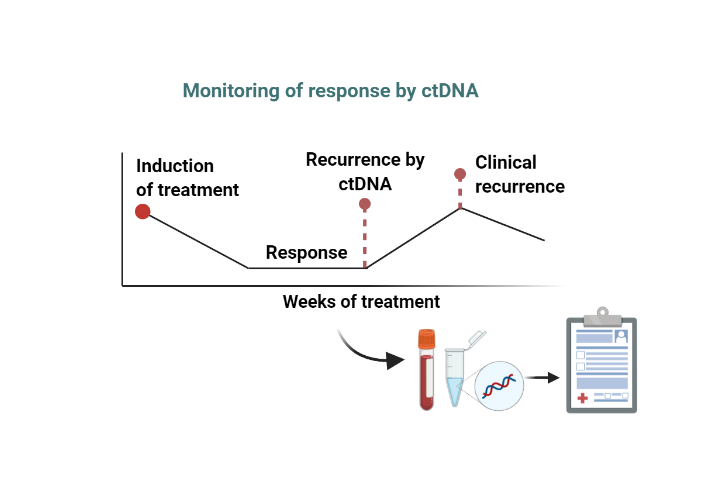
Liquid biopsies offer the promise of earlier detection and monitoring of cancer patients’ treatment progression. However, as applications increasingly rely on ultra low variant allele frequency detection, NGS-based assays are beset with significantly more noise below 0.5% VAF that increases the rate of false positives. Laboratories looking to implement ctDNA assays need better quality controls that can ensure accurate detection and reporting by variant calling algorithms.Standardizing within patient measurements and across sites and platforms are equally critical to ensuring accurate reporting of results and harmonization of patient care.
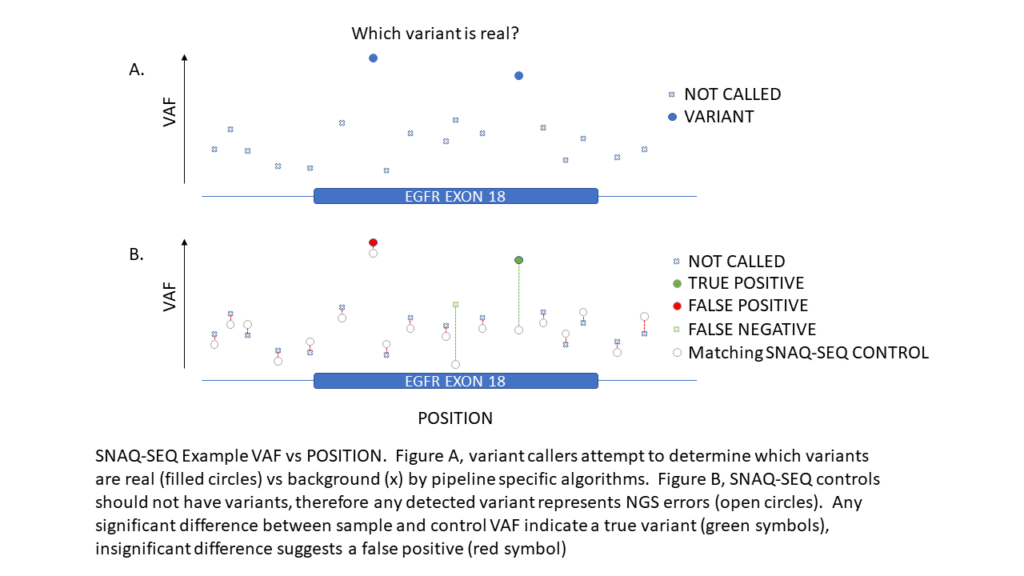
SNAQ-SEQ spike-in standards are highly multiplexed mixtures of synthetic DNA constructs to targeted regions of highest clinical importance. They are formulated to a size profile of ~160bp, approximating cfDNA in form and function to provide high confidence calls for all regions of interest. SNAQ-SEQ standards are spiked into EVERY sample and measure background errors due to systemic (eg. assay, sequencer, and variant caller) or technical (operator, reagents, and consumables) errors and are able to eliminate false positive variant calls due to these factors.
Monitoring minimum residual disease (MRD) requires the highest sensitivity and confidence that a negative result is due to the continued suppression of disease vs. assay performance drift or failure. SNAQ-SEQ can provide limit of detection (LOD) assurance for EVERY sample to assure the assay met its performance specifications, even in absence of target.
Additional Resources:
Presentations:

