Copy Number Variants (CNV) are a clinically important variant type to identify in both somatic and germline genomes. NGS workflows and analytics are increasingly able to resolve duplications, deletions, and other CNVs, alongside SNVs and Indels in a streamlined workflow. These detection algorithms rely on consistent coverage and reads across the genome to accurately identify copy number changes within the genome. However, GC rich regions, suboptimal capture and sequencing primers can impact resolution of CNVs and result in ‘blind spots’ and potentially missed calls.
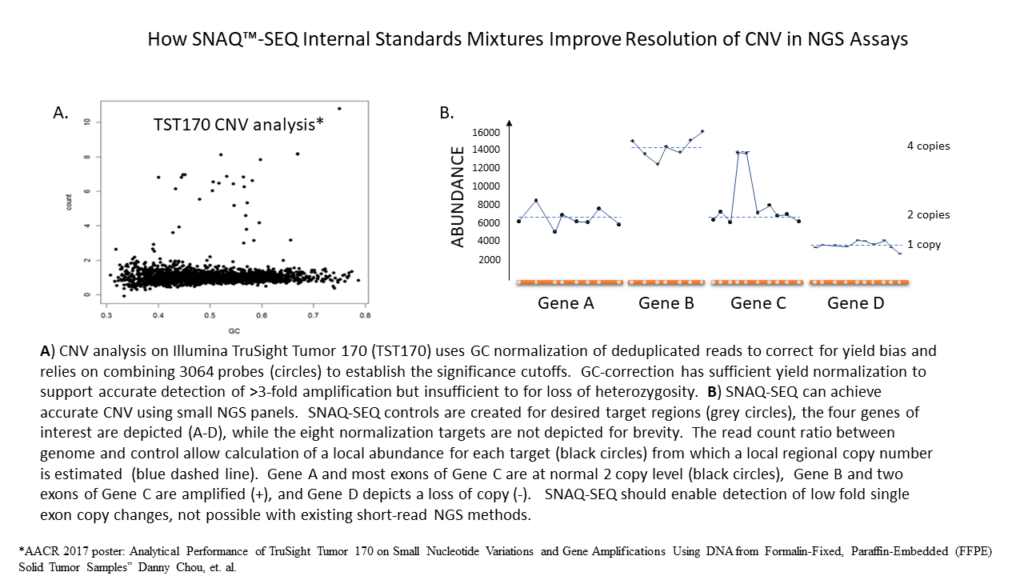
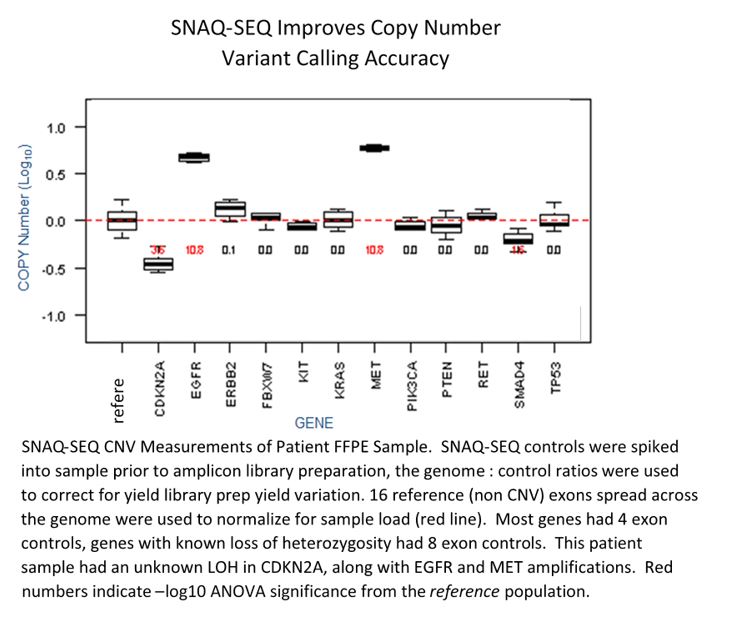
SNAQ-SEQ multiplexed DNA standards can significantly improve the call accuracy and resolution of CNVs by measuring and adjusting for read bias around target genes due to their homology and being added directly to every sample. The result is CNV detection approaching dPCR-like sensitivity—able to resolve from as little as +3 copies to -0.5 copies across a range of alteration sizes.